Endogenous flavivirid (EFV) data
We have used GLUE to organise the 'genomic fossil record' of flavivirids. This page provides a description of Flavivirid-GLUE's paleovirus component, and quick links to specific data items.
Please note: links to files on GitHub are mainly designed to indicate where these files are located within the repository. To investigate files (e.g. tree files) in the appropriate software context we recommend downloading the entire repository and browsing locally.
How were the EFV data generated?
EFV sequences were recovered from whole genome sequence (WGS) assemblies via database-integrated genome screening (DIGS) using the DIGS tool.
All data pertaining to this screen are included in this repository.
- The complete list of vertebrate genomes screened can be found here.
- The complete list of invertebrate genomes screened can be found here.
- The set of flavivirus polypeptide sequences used as probes can be found here.
- The final set of flavivirus and EFV polypeptide sequences used as references can be found here.
- Input parameters for screening using the DIGS tool can be found here.
Nomenclature for EFVs
We have applied a systematic approach to naming EFV, following a convention developed for endogenous retroviruses. Each element was assigned a unique identifier (ID) constructed from a defined set of components.

The first component is the classifier ‘EFV’ (endogenous flavivirid).
The second component is a composite of two distinct subcomponents separated by a period: (i) the name of EFV group; (ii) a numeric ID that uniquely identifies the insertion. The numeric ID is an integer that identifies a unique insertion locus that arose as a consequence of an initial germline infection. Thus, orthologous copies in different species are given the same number.
The third component of the ID defines the set of host species in which the ortholog occurs.
Paleovirus-specific schema extensions
The paleovirus component of Flavivirid-GLUE extends GLUE's core schema to allow the capture of EFV-specific data. These schema extensions are defined in this file and comprise two additional table: 'locus_data' and 'refcon_data'. Both tables are linked to the main 'sequence' table via the 'sequenceID' field.
The 'locus_data' table contains EFV locus information: e.g. species, assembly, scaffold, location coordinates.
The 'refcon_data' table contains summary information for individual EFV insertions. It refers to the reference sequences constructed to represent each insertion, which reflect our best efforts to reconstruct progenitor virus sequences as they might have looked when they initially integrated into the germline of ancestral species.
Raw EFV sequences and data
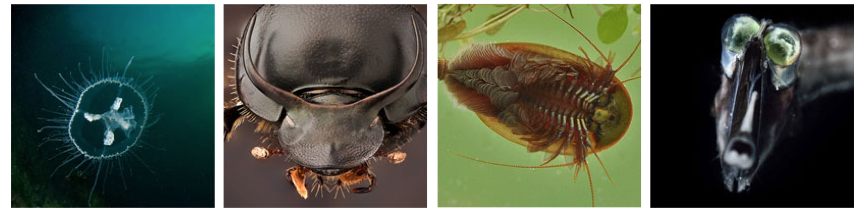
Some of the species in which we identified novel endogenous flaviviral elements (EFVs) Left to right: freshwater jellyfish (Craspedacusta sowerbyi), long-horned beetle (Anoplophora glabripennis), tadpole shrimp (Lepidurus arcticus), tube-eye fish (Stylephorus chordatus).
Raw FASTA for EFVs recovered via database-integrated genome screening (DIGS) are here.
Sequence-associated data in tabular format are here. The tabular files contain information about the genomic locations of EFVs.
EFV reference sequences and data
We constructed reference sequences for EFVs using alignments of EFV sequences derived from the same initial germline colonisation event - i.e. orthologous elements in distinct species, and paralogous elements that have arisen via intragenomic duplication of EFV sequences.
EFV consensus/reference FASTA is here.
Tabular formatted metadata for EFV reference sequences is here.
Phylogenetic trees
We used GLUE to implement an automated process for deriving midpoint rooted, annotated trees from the alignments included in our project.
Trees were constructed at distinct taxonomic levels:
- Major lineage-level phylogenies
- Minor lineage-level phylogenies
- Genus-level phylogenies
- Subgenus-level phylogenies
Related Publications
Bamford CGG, de Souza WM, Parry R and RJ Gifford
(2021)
Comparative analysis of genome-encoded viral sequences reveals the evolutionary history of the Flaviviridae.
preprint [view]
Singer JB, Thomson EC, McLauchlan J, Hughes J, and RJ Gifford
(2018)
GLUE: A flexible software system for virus sequence data.
BMC Bioinformatics
[view]
Zhu H, Dennis T, Hughes J, and RJ Gifford
(2018)
Database-integrated genome screening (DIGS): exploring genomes heuristically using sequence similarity search tools and a relational database.
[preprint]
Gifford RJ, Blomberg B, Coffin JM, Fan H, Heidmann T, Mayer J, Stoye J, Tristem M, and WE Johnson
(2018)
Nomenclature for endogenous retrovirus (ERV) loci.
Retrovirology
[view]