The alignment tree in Hepadnaviridae-GLUE
GLUE projects have the option of using a data structure called an alignment tree to link constrained multiple sequence alignments representing different taxonomic levels, and we've used this approach in Hepadnaviridae-GLUE.
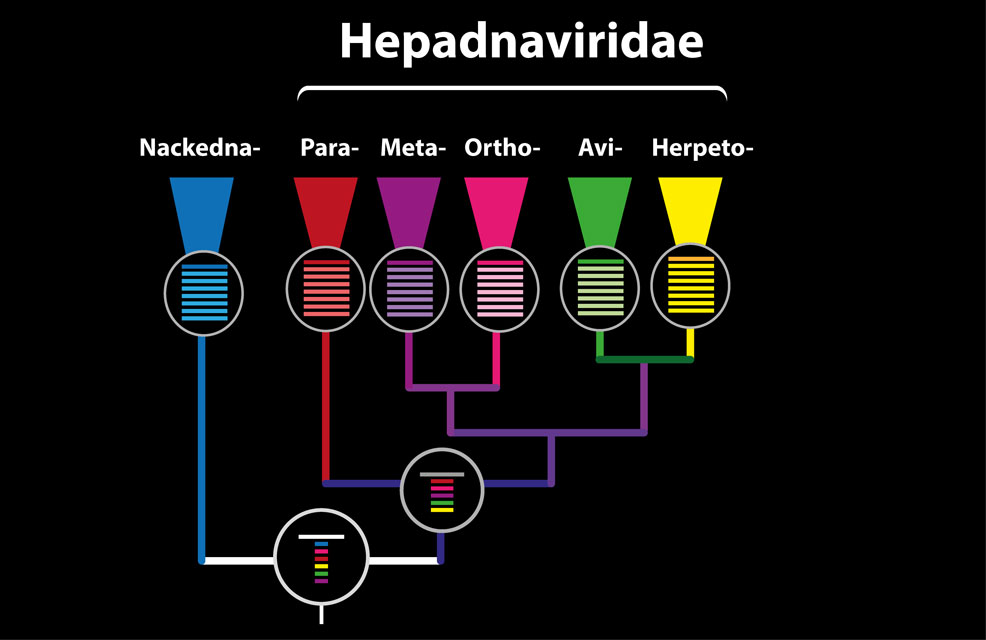
The schematic figure above shows the alignment tree structure in Hepadnaviridae-GLUE. We have constructed 'tip' alignments at genus level, as well as a family-level alignment representing the Hepadnaviridae (located at an internal node in the tree above), and a root alignment that includes the recently described 'nackednaviruses' as an outgroup.
For the lower taxonomic levels (i.e. within and below genus level) we aligned complete coding sequences. For the highest taxonomic levels (i.e. at the root) we aligned only the most conserved gene (the viral polymerase). We used an alignment tree data structure to link these alignments, via a set of common reference sequences. The root alignment contains all reference sequences, whereas all children of the root inherit at least one reference from their immediate parent. Thus, all alignments are linked to one another via our chosen set of master reference sequences.
The example below illustrates some of the advantages of this. The node representing the root of the Hepadnaviridae contains only the master reference sequences for each genus (i.e. it only has five sequences in it). This makes it very easy to maintain, but what if we want to extract an alignment or build a tree at family level that includes all taxa?
We can use the alignment tree to accomplish this, as shown below. On the GLUE console, first let's list the members of the relevant alignment:
Mode path: /
GLUE> project hepadnaviridae alignment AL_Hepadnaviridae list member
+===================+======================+=====================+
| alignment.name | sequence.source.name | sequence.sequenceID |
+===================+======================+=====================+
| AL_Hepadnaviridae | ncbi-refseqs | NC_001344 |
| AL_Hepadnaviridae | ncbi-refseqs | NC_003977 |
| AL_Hepadnaviridae | ncbi-refseqs | NC_027922 |
| AL_Hepadnaviridae | ncbi-refseqs | NC_030445 |
| AL_Hepadnaviridae | ncbi-refseqs | NC_030446 |
+===================+======================+=====================+
AlignmentMembers found: 5
As expected, there are only five members. Now lets look at the AL_Hepadnaviridae alignment to see how it is linked to the other alignments, using the 'list children' command.
Mode path: /
GLUE> project hepadnaviridae alignment AL_Hepadnaviridae list children
+========================+==========================+
| name | refSequence.name |
+========================+==========================+
| AL_Avihepadnavirus | REF_Avi_MASTER_DHBV |
| AL_Herpetohepadnavirus | REF_Herpeto_MASTER_tfHBV |
| AL_Metahepadnavirus | REF_Meta_MASTER_bgHBV |
| AL_Orthohepadnavirus | REF_Ortho_MASTER_HBV |
| AL_Parahepadnavirus | REF_Para_MASTER_wsHBV |
+========================+==========================+
Alignments found: 5
The result shows that - as expected based on the figure above - the Hepadnaviridae alignment is linked to five 'child' alignments, each of which represents a hepadnavirus genus. The constraining reference sequence of each alignment is shown in the table.
Because the alignments are linked, we can use GLUE's fastAlignmentExporter module to link across all of the alignments and export a codon-level alignment that contains all taxa, as follows:
GLUE> project hepadnaviridae module fastaAlignmentExporter
OK
Mode path: /project/hepadnaviridae/module/fastaAlignmentExporter
GLUE> export AL_Hepadnaviridae -r REF_Ortho_MASTER_HBV -f Polymerase -a -e -c -p
A few things to explain about this command:
- The -r option is used to specify a reference sequence (here we use the project master reference, HBV).
- The -f option is used to select a feature within the specified reference (here we select the polymerase gene reading frame).
- The -a option means 'all taxa' - it is provided instead of a where clause (specified using -w option).
- The -e option means 'exclude empty rows' - it prevents inclusion of taxa that do not span the selected feature in the exported alignment.
- The -c option is the important one for our example - it means 'export recursively' - in other words, include taxa in alignments that are children of the specified alignment.
- The -p option is means 'preview on the command line' - this will output the result of the command to the console. To export the alignment as a file use the -o option with a file name, as shown here:
Mode path: /project/hepadnaviridae/module/fastaAlignmentExporter
GLUE> export AL_Hepadnaviridae -r REF_Ortho_MASTER_HBV -f Polymerase -a -e -c -o out.fasta